Project Focus
Project Leaders
1. Joseph Picone, PhD; Iyad Obeid, PhD
Neural Engineering Data Consortium
College of Engineering, Temple University
Philadelphia, Pennsylvania, U.S.A.
2. Sanda M. Harabagiu, PhD
Human Language Technology Research Institute
University of Texas at Dallas
Dallas, Texas, U.S.A.
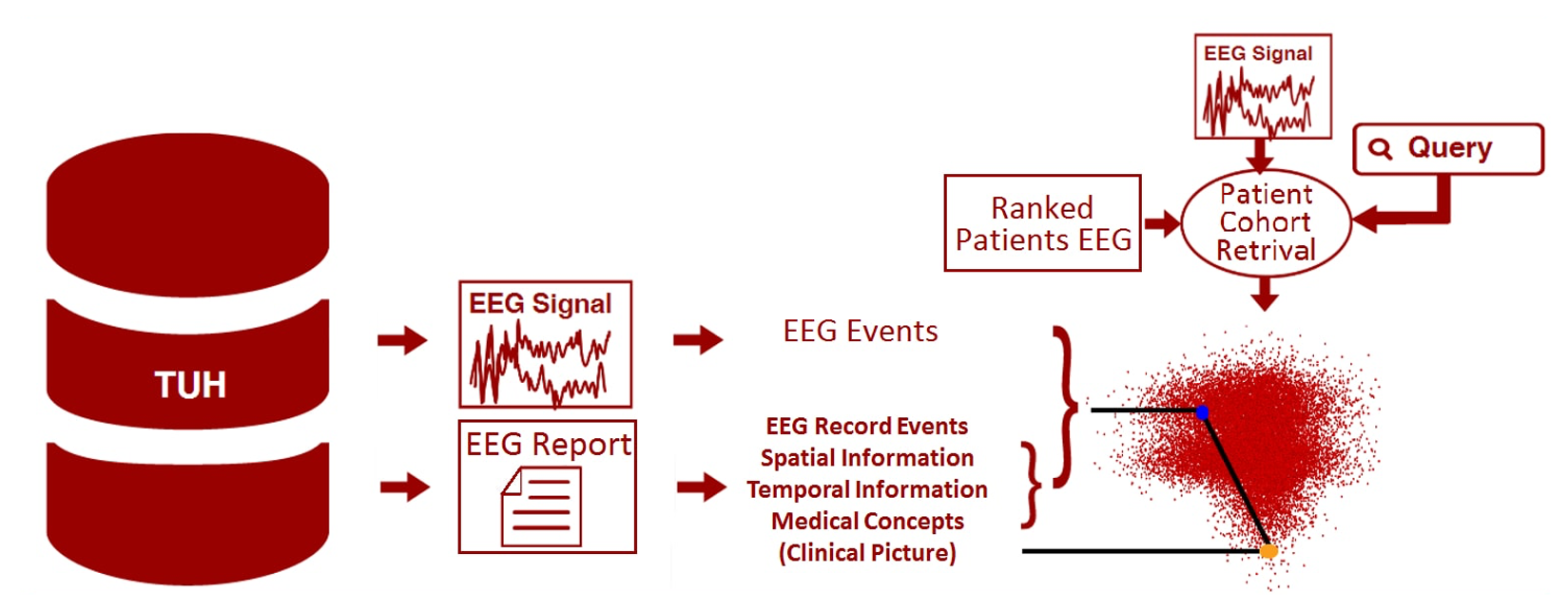
Electronic medical records (EMRs) collected at every hospital in the country collectively contain a staggering wealth of biomedical knowledge. EMRs can include unstructured text, temporally constrained measurements (e.g., vital signs), multichannel signal data (e.g., EEGs), and image data (e.g., MRIs). This information could be transformative if properly harnessed. Information about patient medical problems, treatments, and clinical course is essential for conducting comparative effectiveness research. Uncovering clinical knowledge that enables comparative research is the primary goal of this research.
Our focus in this research project is the automatic interpretation of a
clinical EEG big data resource known as the TUH EEG Corpus (TUH EEG).
This corpus was collected over 14 years at Temple University Hospital
and consists of over 28,000 sessions and 15,000 patients. Clinicians will
be able to retrieve relevant EEG signals and EEG reports using standard
queries (e.g. “Young patients with focal cerebral dysfunction who were
treated with Topamax”). We will automatically annotate EEG events that
contribute to a diagnosis. Automated techniques are used to discover and
time-align the underlying EEG events using semi-supervised learning.
Clinical concepts, their type, polarity and modality are being discovered
automatically, as well as spatial and temporal information. In addition,
we are extracting the medical concepts describing the clinical picture
of patients from the EEG reports. We are developing a patient cohort
retrieval system that will operate on the extracted clinical knowledge.
An important outcome of this research will be the existence of an
annotated big data archive of EEGs that will greatly increase
accessibility for non-experts in neuroscience, bioengineering and medical
informatics who would like to study EEG data. The creation of this
resource through the development of efficient automated data wrangling
techniques will demonstrate that a much wider range of big data
bioengineering applications are now tractable.
